 Insect and spider predators found in arable crops are known to consume crop pests, but also other invertebrates, known as alternative prey. These generalist natural enemies can attain high densities by feeding on alternative prey during the early growing season, before any pest outbreak occurs. This could be useful for pest control before specialist natural enemies, like parasitic wasps, appear.
Insect and spider predators found in arable crops are known to consume crop pests, but also other invertebrates, known as alternative prey. These generalist natural enemies can attain high densities by feeding on alternative prey during the early growing season, before any pest outbreak occurs. This could be useful for pest control before specialist natural enemies, like parasitic wasps, appear.
However, the alternative prey on which the generalist natural enemies feed also includes some species that are also pest natural enemies. This preliminary work looks at predation on money spiders (Linyphiidae), known to be effective aphid predators, by a ground beetle, Pterostichus melanarius, which consumes both slugs and aphids. We wanted to establish whether these beetles consumed any of these spiders and, if so, whether they show a preference for those species who also occur in the same places as the beetles.
Figure 1: Percentage of 622 ground beetles Pterostichus melanarius testing positive (PCR) for the consumption of four common spider taxa in winter wheat during summer 2006
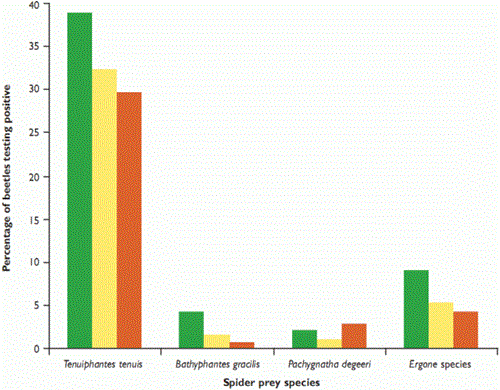
Revealing patterns of predation among invertebrates is difficult. Analysis of gut contents is useful, but can be biased owing to differing levels of expertise among researchers and the fact that only hard parts of the prey can be detected. Our work relies on a technique called polymerase chain reaction (PCR), which allows prey DNA to be recognised in the gut contents of the predators. PCR is explained in the box. Over June, July and August 2006, we caught a total of 622 P melanarius beetles using pitfall traps placed in a 16m x 16m grid in a field of winter wheat at Rothamsted Research Institute, Harpenden. We then removed the beetles’ guts and extracted DNA.
 We then screened this DNA with prey-specific molecular markers (PCR primers) to determine whether they had eaten the money spiders Tenuiphantes tenuis, Bathyphantes gracilis and Erigone species, as well as the tetragnathiid spider Pachygnatha degeeri. The sensitivity of the prey detection tests was calibrated using controlled feeding trials. We fed each beetle predator a single individual of the target spider species, after which we killed batches by freezing at regular time intervals following prey consumption. We screened DNA extracted from the guts of these beetles using PCR. We measured the rate of decay in detectability for each prey DNA as the time after prey consumption at which 50% tested positive. When a population of predators is screened, this can be used to weight the estimate (because the time since they last ate will vary) of the proportion of predators consuming each prey species.
We then screened this DNA with prey-specific molecular markers (PCR primers) to determine whether they had eaten the money spiders Tenuiphantes tenuis, Bathyphantes gracilis and Erigone species, as well as the tetragnathiid spider Pachygnatha degeeri. The sensitivity of the prey detection tests was calibrated using controlled feeding trials. We fed each beetle predator a single individual of the target spider species, after which we killed batches by freezing at regular time intervals following prey consumption. We screened DNA extracted from the guts of these beetles using PCR. We measured the rate of decay in detectability for each prey DNA as the time after prey consumption at which 50% tested positive. When a population of predators is screened, this can be used to weight the estimate (because the time since they last ate will vary) of the proportion of predators consuming each prey species.
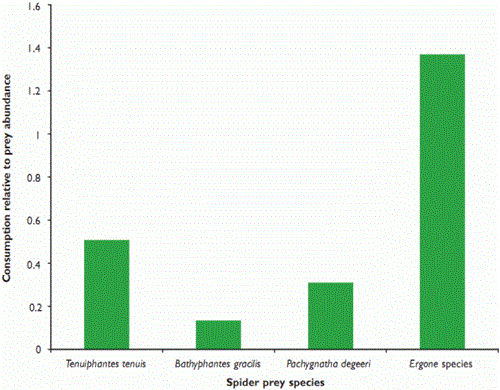
We expected that spiders which lived closer to the ground would suffer from higher rates of predation by P. melanarius, and found that that this was generally the case. Erigone species spin their webs at ground level, whereas T. tenuis and B. gracilis attach theirs to the wheat stems, at around 36mm and 45mm above the ground, respectively. Pachygnatha degeeri hunts, without a web, in the crop foliage. T. tenuis was the spider most often detected (see Figure 1), but it was also the most common, so the impact of predation on it may not be as marked as in some other taxa, particularly the Erigone species (see Figure 2). We will continue to investigate predation rates of different prey types by modelling encounter rates.
Figure 2: Consumption of four spider taxa by the beetle Pterostichus melanarius in relation to relative prey availability in winter wheat during June 2006
An explanation of PCR
PCR is a process in which, firstly, DNA is heated so that its two complementary strands dissociate. The reagents are then cooled to such a degree that a pair of primers – sequences of single-stranded DNA, 20-30 base pairs long – bind to either end of a short, unique region of the genome. The temperature is then raised again to a level suitable for an enzyme, DNA polymerase, to extend a new copy of the target sequence. Cycles of these temperature changes doubles the amount of the target sequence, producing sufficient copies of a short, unique DNA sequence to visualise under UV light.
Acknowledgements
This work was carried out by Jeff Davey as part of his NERC/GWCT CASE PhD studentship supervised at Cardiff University by Bill Symondson and John Holland from the GWCT. Predator samples were collected as part of a larger BBSRC-funded project between Bill Symondson at Cardiff and David Bohan at Rothamsted Research (Dynamic responses of predators to biodiversity in sustainable agriculture: spatial and molecular analyses, BB/D001188/1).